Computational and Quantitative Biology PhD
Francesco Iorio - CRISPR-cas9 screens and multi-omic data integration for identifying and prioritising anti-cancer therapeutic targets
Venue: Teams
ABSTRACT
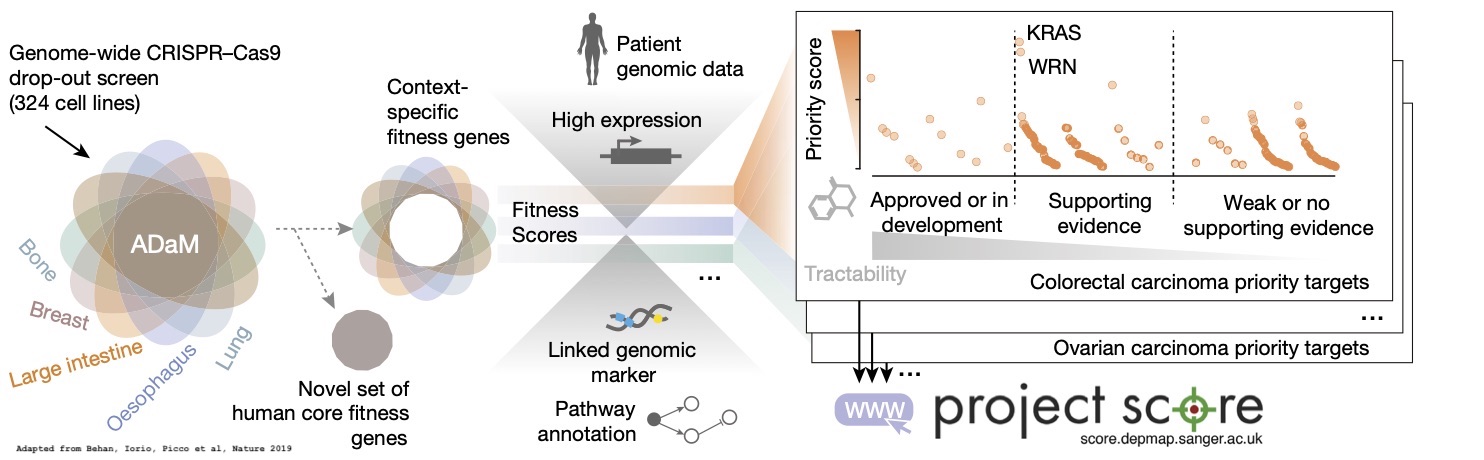
Functional genomics approaches can overcome limitations—such as the lack of identification of robust targets and poor clinical efficacy—that hamper cancer drug development. We performed genome-scale CRISPR–Cas9 screens in 324 human cancer cell lines from 30 cancer types and developed a data-driven framework to prioritize candidates for cancer therapeutics. We integrated cell fitness effects with genomic biomarkers and target tractability for drug development to systematically prioritize new targets in defined tissues and genotypes. We verified one of our most promising dependencies, the Werner syndrome ATP-dependent helicase, as a synthetic lethal target in tumours from multiple cancer types with microsatellite instability. Our analysis provides a resource of cancer dependencies, generates a framework to prioritize cancer drug targets and suggests specific new targets. The principles described in this study can inform the initial stages of drug development by contributing to a new, diverse and more effective portfolio of cancer drug targets
SPEAKER
Francesco Iorio is a Research Group Leader in Computational Biology at the Human Technopole (Milan, Italy) where he is establishing a research group in Computational Pharmacogenomics and Therapeutic Target Discovery. He has been awarded a joint EMBL – European Bioinformatics Institute (EBI) and the Wellcome Sanger Institute (WSI) post-doctoral (ESPOD) fellowship. Following this, He worked as senior bioinformatician at EBI and currently leads the Cancer Dependency Map Analytics team at the WSI.
HOW TO ATTEND
The seminar will be online using TEAMS. Please register to the seminar. We will send you an invite with the link to the seminar.
All Dates
- 2020-12-04 15:00
Powered by iCagenda





